Control flow graphs (CFGs) and function call graphs (FCGs) have become pivotal in providing a detailed understanding of program execution and effectively characterizing the behaviour of malware. These graph-based representations, when combined with graph neural networks, have shown promise in developing high-performance malware detectors.
As part of our work, we generate CFGs and FCGs for the BODMAS, DikeDataset, and pe-machine-learning-dataset datasets using the angr Python library. Additionally, we also provide embeddings of graphs and explanations for use in machine learning tasks. Below is an example of the pipeline used to generate the graphs from our work.
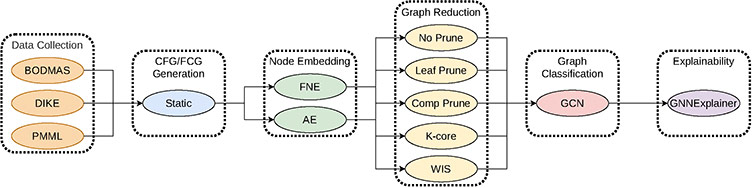
Furthermore, this contributes to the field of towards the task of graph classification on large graphs with many samples in the field of graph learning. Common datasets for this task consist of many samples with few hundred nodes on average, or few samples with few thousand and many classes. This work proposes many graphs of large size, some with hundreds of thousands of nodes and edges, and two classes, malicious and benign.
We recognize two main audiences for this work: one, researchers in the field of malware detection and analysis, and two, researchers in the field of graph-based machine learning. The former may be interested in the Attribute Graphs whereas the latter may be interested in the Embedded and Explained Graphs, all of which are described below.
Importantly, all samples with the same filename, excluding the file extension, originate from the same binary file. Derived from the sha256 hash, depending on if the original file required arming or not. However, only files generated from the same underlying lineage are comparable. For example, a file in exps/AE/DikeDataset is only comparable with a file in ebds/AE/DikeDataset and the corresponding graph type, in this case CFG, within the file in cfgs_fcgs/DikeDataset.
Please refer to the src directory for an example of how to start working with the dataset in index.py. We refer users to angr, PyTorch Geometric, and NetworkX for further information on working with the underlying dataset objects and libraries.
We are fully aware of the presence of isomorphic samples within the dataset. We knowingly include these samples, not only for completeness, but importantly because the sample graphs, while isomorphic, do not originate from "true" duplicates with respect to the original binary samples. We leave it to the end user to decide how to handle such samples. We understand that removing isomorphic graphs may be of particular interest for graph-based machine learning whereas in malware analysis it may not be a concern.
One general approach to know which samples are isomorphic, with high probability, is simply to compare the number of nodes, edges, and components in the graphs based on the provided CSV files and then test for isomorphism.
There are many speculative reasons for the presence of isomorphic samples. Presumably, malware authors may alter source code, or the binary itself, to perturb its signature and evade detection while also leaving the underlying CFG/FCG intact. Additionally, some samples may originate from the same malware family and thus have the same CFG/FCG.
YouTube video: "CIC Statically Generated Graphs for Malware Analysis (CIC-SGG-2024)" by Griffin Higgins, Cybersecurity Software Developer, Canadian Institute for Cybersecurity with introduction and Q&A by Sumit Kundu.
L. Yang, A. Ciptadi, I. Laziuk, A. Ahmadzadeh, and G. Wang, “Bodmas: An open dataset for learning based temporal analysis of pe malware,” in 2021 IEEE Security and Privacy Workshops (SPW), pp. 78–84, IEEE, 2021.
G.-A. Iosif, “Dikedataset,” 2021. Accessed on February 27, 2024.
Practical Security Analytics LLC, “Pe malware machine learning dataset,” 2024. Accessed: 2024-08-06.
The CIC-SGG-2024 dataset is publicly available for researchers. If you are using our dataset, you must cite our related research paper that covers important details related to its usage and application.
H. Mohammadian, G. Higgins, S. Ansong, R. Razavi-Far, A. Ghorbani. "Explainable Malware Detection through Integrated Graph Reduction and Learning Techniques," Big Data Research, Aug 2025.
Curated by Griffin Higgins, please direct questions to griffin.higgins@unb.ca. Only questions with the heading subject "CIC-SGG-2024" will be guaranteed to receive a reply.